Figure 17.3.2 Real-time detection capability of the Antibody Beacon Tyrosine Kinase Assay Kit (A35725). Fluorescence of the Antibody Beacon detection complex in tyrosine kinase assay buffer was monitored over time. After ~15 seconds, an excess of phosphotyrosine-containing peptide was added to the Antibody Beacon detection complex and the off-rate was calculated.
Page Contents
The cascade of cellular events in response to an internal signal or environmental stimulus requires a diversity of molecular participants, ranging from ions to enzymes. Signal transduction pathways frequently activate specific protein kinases, leading to the phosphorylation of particular cellular proteins and subsequent initiation of a multitude of cellular responses. Binding and hydrolysis of nucleotides plays a major role in these activities, and our nucleotide analogs and assays for phosphate-producing enzymes are important tools for signal transduction research and high-throughput screening of compounds that affect signal transduction.
We offer a selection of native and modified biomolecules to aid the researcher in dissecting this highly complex branch of the signal transduction process. In addition to the probes below, we have developed the PiPer and EnzChek assay kits for quantitation of inorganic phosphate and pyrophosphate that are extremely useful for following hydrolysis of nucleotides by various enzymes and of phosphate esters by protein phosphatases. These kits and other kits to measure ATP by chemiluminescence and protein phosphatase activity are described in Detecting Enzymes That Metabolize Phosphates and Polyphosphates—Section 10.3.
Protein kinases are critical players in signal transduction pathways. The fluorometric assay of kinases, however, is not straightforward because ATP-dependent phosphorylation of a fluorescent peptide substrate does not directly lead to appreciable changes in the fluorescence of the product.![]() We provide an extensive range of assays for protein kinases that utilize a variety of strategies to detect phosphorylation of peptide and protein substrates (Invitrogen kinase assay platforms—Table 17.1).
We provide an extensive range of assays for protein kinases that utilize a variety of strategies to detect phosphorylation of peptide and protein substrates (Invitrogen kinase assay platforms—Table 17.1).
Antibody Beacon Tyrosine Kinase Assay Kit
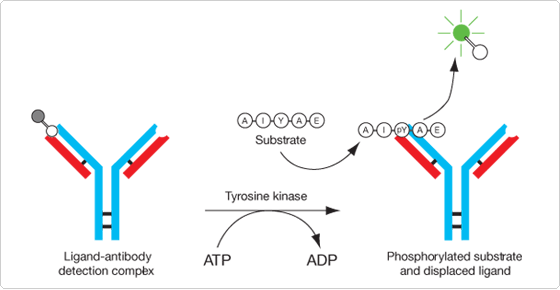
The Antibody Beacon Tyrosine Kinase Assay Kit (A35725) provides a homogeneous solution assay for measuring the activity of tyrosine kinases and the effectiveness of potential inhibitors and modulators.![]() The key to this tyrosine kinase assay is a small-molecule tracer ligand labeled with our bright green-fluorescent Oregon Green 488 dye. When an anti-phosphotyrosine antibody binds this tracer ligand to form the Antibody Beacon detection complex, the fluorescence of the Oregon Green 488 dye is efficiently quenched. In the presence of a phosphotyrosine-containing peptide, however, this Antibody Beacon detection complex is rapidly disrupted, releasing the tracer ligand and relieving its antibody-induced quenching (Figure 17.3.1). Upon its displacement by a phosphotyrosine residue, the Oregon Green 488 dye–labeled tracer ligand exhibits an approximately fourfold fluorescence enhancement, enabling the detection of as little as 50 nM phosphotyrosine-containing peptide with excellent signal-to-background discrimination. Key benefits of the Antibody Beacon Tyrosine Kinase Assay Kit include:
The key to this tyrosine kinase assay is a small-molecule tracer ligand labeled with our bright green-fluorescent Oregon Green 488 dye. When an anti-phosphotyrosine antibody binds this tracer ligand to form the Antibody Beacon detection complex, the fluorescence of the Oregon Green 488 dye is efficiently quenched. In the presence of a phosphotyrosine-containing peptide, however, this Antibody Beacon detection complex is rapidly disrupted, releasing the tracer ligand and relieving its antibody-induced quenching (Figure 17.3.1). Upon its displacement by a phosphotyrosine residue, the Oregon Green 488 dye–labeled tracer ligand exhibits an approximately fourfold fluorescence enhancement, enabling the detection of as little as 50 nM phosphotyrosine-containing peptide with excellent signal-to-background discrimination. Key benefits of the Antibody Beacon Tyrosine Kinase Assay Kit include:
- Real-time measurements. Unlike many other commercially available tyrosine kinase assays, the Antibody Beacon Tyrosine Kinase Assay Kit permits real-time monitoring of kinase activity (Figure 17.3.2). Not only is the Antibody Beacon detection complex rapidly dissociated in the presence of phosphotyrosine residues, but the assay components have been designed to be simultaneously combined, eliminating any delay in the measurements.
- Simple detection protocol. Tyrosine kinase activity is measured by a simple increase in fluorescence intensity; no special equipment, additional reagents, or extra steps are required. This assay is readily compatible with any fluorescence microplate reader.
- Use of natural substrates. The Antibody Beacon tyrosine kinase assay utilizes unlabeled peptide or protein substrates, is compatible with substrates that are pre-phosphorylated at serine or threonine (but not at tyrosine) residues and is applicable to the assay of a wide variety of kinases.
- Compatibility. The anti-phosphotyrosine antibody provided in the Antibody Beacon Tyrosine Kinase Assay Kit is specific for phosphotyrosine residues; assay components such as ATP (up to 1 mM) and reducing agents such as dithiothreitol (DTT, up to 1 mM) do not interfere with this assay. This anti-phosphotyrosine antibody was selected from among several clones to produce the greatest fluorescence enhancement by the kinase-phosphorylated product.
- Reliability. This tyrosine kinase assay has a broad signal window, indicated by a Z' factor
of >0.85.
The Antibody Beacon Tyrosine Kinase Assay Kit comes with all the reagents needed to perform this assay, including:
- Oregon Green 488 dye–labeled tracer ligand
- Anti-phosphotyrosine antibody
- Concentrated tyrosine kinase reaction buffer
- Two generic tyrosine kinase substrate solutions: a poly(Glu:Tyr) solution and a poly(Glu:Ala:Tyr) solution
- Dithiothreitol (DTT)
- Adenosine triphosphate (ATP)
- Phosphotyrosine-containing peptide, phospho-pp60 c-src (521–533), for use as a reference
- Detailed protocols (Antibody Beacon Tyrosine Kinase Assay Kit)
Each kit provides sufficient reagents to perform ~400 assays using a 50 µL assay volume in a fluorescence microplate reader.

Fluorescent Polymyxin B Analogs
Polymyxin B is a cyclic polycationic peptide antibiotic (Figure 17.3.3) that binds to lipopolysaccharides and anionic lipids.![]() Polymyxin B is also a selective inhibitor of protein kinase C, with an IC50 of ~35 µM,
Polymyxin B is also a selective inhibitor of protein kinase C, with an IC50 of ~35 µM,![]() as well as a potent inhibitor of calmodulin, with an IC50 of 80 nM in the presence of 500 µM Ca2+.
as well as a potent inhibitor of calmodulin, with an IC50 of 80 nM in the presence of 500 µM Ca2+.![]() Our fluorescent polymyxin B analogs include those of the green-fluorescent BODIPY FL
Our fluorescent polymyxin B analogs include those of the green-fluorescent BODIPY FL ![]() and Oregon Green 514 fluorophores (P13235, P13236), as well as the ultraviolet light–excitable dansyl polymyxin
and Oregon Green 514 fluorophores (P13235, P13236), as well as the ultraviolet light–excitable dansyl polymyxin ![]() (P13238).
(P13238).

Figure 17.3.3. Structure of polymyxin B.
Hypericin
Hypericin (H7476), an anthraquinone derivative isolated from plants of the genus Hypericum,![]() is a potent, selective inhibitor of PKC (IC50 = 1.7 µg/mL = 3.4 µM) useful for probing and manipulating PKC in live cells.
is a potent, selective inhibitor of PKC (IC50 = 1.7 µg/mL = 3.4 µM) useful for probing and manipulating PKC in live cells.![]() Hypericin has a variety of pharmacological properties, from antibacterial and antineoplastic activities to antiviral activities
Hypericin has a variety of pharmacological properties, from antibacterial and antineoplastic activities to antiviral activities ![]() and induction of apoptosis.
and induction of apoptosis.![]() Hypericin is also a potent photosensitizer, with a quantum yield of 0.75 for the generation of singlet oxygen.
Hypericin is also a potent photosensitizer, with a quantum yield of 0.75 for the generation of singlet oxygen.![]()
RediPlate 96 EnzChek Tyrosine Phosphatase Assay Kits
Protein tyrosine phosphatases (PTP) represent a large family of enzymes that play a very important role in intra- and intercellular signaling. PTPs work antagonistically with protein tyrosine kinases to regulate signal transduction pathways in response to a variety of signals, including hormones and mitogens.![]() Our RediPlate 96 EnzChek Tyrosine Phosphatase Assay Kit (R22067) provide researchers with a sensitive and convenient method to monitor PTP and screen PTP inhibitors in a variety of research areas
Our RediPlate 96 EnzChek Tyrosine Phosphatase Assay Kit (R22067) provide researchers with a sensitive and convenient method to monitor PTP and screen PTP inhibitors in a variety of research areas ![]() (Figure 17.3.4).
(Figure 17.3.4).
The EnzChek tyrosine phosphatase assay is based on 6,8-difluoro-4-methylumbelliferyl phosphate ![]() (DiFMUP, D6567, D22065; Detecting Enzymes That Metabolize Phosphates and Polyphosphates—Section 10.3). Unlike other end-point tyrosine phosphatase assay kits, the EnzChek tyrosine phosphatase assay is continuous, allowing researchers to easily measure fluorescence at various time points in order to follow the kinetics of the reaction. Furthermore, the assay is not affected by free phosphate and is compatible with most nonionic detergents, minimizing sample processing before analysis. Most importantly, each assay well contains inhibitors to help ensure that the assay is selective for tyrosine phosphatases; other phosphatases, including serine/threonine phosphatases, will not hydrolyze DiFMUP under our assay conditions (Figure 17.3.5). Unlike phosphopeptide-based assays, this DiFMUP-based assay can be used to monitor a variety of tyrosine phosphatases, including PTP-1B and CD-45 (Figure 17.3.5). Tyrosine phosphatase inhibitors can be evaluated quantitatively in the assay for their effect on tyrosine phosphatase activity.
(DiFMUP, D6567, D22065; Detecting Enzymes That Metabolize Phosphates and Polyphosphates—Section 10.3). Unlike other end-point tyrosine phosphatase assay kits, the EnzChek tyrosine phosphatase assay is continuous, allowing researchers to easily measure fluorescence at various time points in order to follow the kinetics of the reaction. Furthermore, the assay is not affected by free phosphate and is compatible with most nonionic detergents, minimizing sample processing before analysis. Most importantly, each assay well contains inhibitors to help ensure that the assay is selective for tyrosine phosphatases; other phosphatases, including serine/threonine phosphatases, will not hydrolyze DiFMUP under our assay conditions (Figure 17.3.5). Unlike phosphopeptide-based assays, this DiFMUP-based assay can be used to monitor a variety of tyrosine phosphatases, including PTP-1B and CD-45 (Figure 17.3.5). Tyrosine phosphatase inhibitors can be evaluated quantitatively in the assay for their effect on tyrosine phosphatase activity.
Each RediPlate 96 EnzChek Tyrosine Phosphatase Assay Kit (R22067) includes:
- One RediPlate 96 EnzChek tyrosine phosphatase assay 96-well microplate
- Reaction buffer
- Detailed assay protocols (RediPlate 96 EnzChek Tyrosine Phosphatase Assay Kit)

RediPlate 96 EnzChek Serine/Threonine Phosphatase Assay Kit
The majority of protein phosphorylation occurs on serine and threonine residues, with <0.01–0.05% on tyrosine residues. Serine/threonine phosphatases represent a large family of enzymes that have been implicated in the regulation of metabolism, transcription, translation, differentiation, cell cycle, cytoskeletal dynamics, oncogenesis and signal transduction. The RediPlate 96 EnzChek Serine/Threonine Phosphatase Assay Kit (R33700) provides a fast, simple and direct fluorescence-based assay for detecting serine/threonine phosphatases and their corresponding modulators and inhibitors ![]() (Figure 17.3.4).
(Figure 17.3.4).
As with the RediPlate 96 EnzChek Tyrosine Phosphatase Kit, the substrate incorporated in the RediPlate 96 EnzChek Serine/Threonine Phosphatase Assay Kit is DiFMUP. Inhibitors are included in each assay well to help ensure that the assay is selective for serine/threonine phosphatases; under the prescribed assay conditions, other phosphatases, including tyrosine phosphatases, do not significantly react with the substrate (Figure 17.3.6). Furthermore, unlike phosphopeptide-based assays, this DiFMUP-based assay can be used to monitor a variety of serine/threonine phosphatases including PP-1, PP-2A and PP-2B (Figure 17.3.6). Serine/threonine phosphatase inhibitors can be evaluated quantitatively in the assay for their effect on serine/threonine phosphatase activity (Figure 17.3.7). Additional advantages of this RediPlate assay include compatibility with nonionic detergents and insensitivity to free phosphate, minimizing sample processing before analysis.
Each RediPlate 96 EnzChek Serine/Threonine Phosphatase Assay Kit includes:
- One RediPlate 96 EnzChek serine/threonine phosphatase assay 96-well microplate
- Concentrated reaction buffer
- NiCl2
- MnCl2
- Dithiothreitol
- Detailed assay protocols (RediPlate 96 EnzChek Serine/Threonine Phosphatase Assay Kit)
To ensure the integrity of the pre-dispensed reagents, the 96-well microplate provided in both RediPlate Protein Phosphatase Assay Kits is packaged in a resealable foil pouch and consists of twelve removable strips, each with eight wells (Figure 17.3.8). Eleven of the strips (88 wells) are preloaded with the fluorogenic substrate DiFMUP; the remaining strip, marked with black tabs, contains a dilution series of the DiFMU reference standard for generating a standard curve.

Figure 17.3.6 Specificity of the RediPlate 96 EnzChek Serine/Threonine Phosphatase Assay Kit (R33700) for serine/threonine phosphatases. The phosphatases listed in the tables were applied at the indicated concentrations to a RediPlate 96 EnzChek serine/threonine phosphatase assay microplate. Reactions were incubated at 37°C. After 1 hour, fluorescence was measured in a fluorescence microplate reader using excitation at 355 ± 20 nm and emission at 460 ± 12.5 nm

Figure 17.3.7 Detection of PP-2A inhibition by okadaic acid using the RediPlate 96 EnzChek Serine/Threonine Phosphatase Assay Kit (R33700). Each reaction contained 50 µM DiFMUP, 10 mU/mL PP-2A and the indicated concentration (log scale) of okadaic acid in reaction buffer containing 50 mM Tris-HCl, 0.1 mM CaCl2, 1 mM NiCl2, 125 µg/mL bovine serum albumin (BSA) and 0.05% Tween 20. Reactions were incubated at 37°C. After 30 minutes, fluorescence was measured in a fluorescence microplate reader using excitation at 355 ± 20 nm and emission at 460 ± 12.5 nm

Figure 17.3.8 A RediPlate 96 microplate
Pro-Q Diamond Phosphoprotein/Phosphopeptide Microarray Stain Kit
The Pro-Q Diamond Phosphoprotein/Phosphopeptide Microarray Stain Kit (P33706) provides a method for selectively staining phosphoproteins or phosphopeptides on microarrays without the use of antibodies or radioactivity. This kit permits direct detection of phosphate groups attached to tyrosine, serine or threonine residues in a microarray environment and has been optimized for microarrays with acrylamide gel surfaces. Each Pro-Q Diamond Phosphoprotein/Phosphopeptide Microarray Stain Kit provides:
- Pro-Q Diamond phosphoprotein/phosphopeptide microarray stain
- Pro-Q Diamond microarray destain solution
- Microarray staining gasket with seal tabs, 10 chambers
- Slide holder tube, 20 tubes
- Detailed protocols (Pro-Q Diamond Phosphoprotein/Phosphopeptide Microarray Stain Kit)
The Pro-Q Diamond Phosphoprotein/Phosphopeptide Microarray Stain Kit is ideal for identifying kinase targets in signal transduction pathways and for phosphoproteomics studies.![]()
3',5'-Cyclic AMP (cAMP) is an important second messenger in many signal transduction pathways, linking activation of cell-surface membrane receptors to intracellular responses, and ultimately, to changes in gene expression. cAMP is synthesized by plasma membrane–bound adenylate cyclase, which is coupled to transmembrane receptors for hormones, neurotransmitters and other signaling molecules by heterotrimeric G-proteins. Upon ligand binding, the intracellular receptor domain of a G-protein–coupled receptor (GPCR) interacts with a G-protein, which then dissociates and activates adenylate cyclase, resulting in an increase in the concentration of intracellular cAMP. Subsequently, cAMP activates cAMP-dependent protein kinases (protein kinase A), which phosphorylate specific substrate proteins, including enzymes, structural proteins, transcription factors and ion channels.
Adenylate Cyclase Probe: BODIPY FL Forskolin
Forskolin, isolated from Coleus forskohlii, is a potent activator of adenylate cyclase, the enzyme that catalyzes the formation of cAMP from ATP. Green-fluorescent BODIPY FL forskolin (B7469) has been used to visualize adenylyl cyclase internalization and subcellular distribution,![]() as well as for the pharmacological characterization of adenylyl cyclase catalytic subunits.
as well as for the pharmacological characterization of adenylyl cyclase catalytic subunits.![]()
cAMP Chemiluminescent Immunoassay Kit
The cAMP Chemiluminescent Immunoassay Kit enables ultrasensitive determination of 3',5'-cyclic AMP (cAMP) levels in cell lysates, providing the highest sensitivity of any commercially available cAMP assay. As few as 60 femtomoles of cAMP can be detected. Furthermore, this assay has a wide dynamic range, detecting from 0.06 to 6000 picomoles without the need for sample dilution or manipulations such as acetylation. This extensive dynamic range is especially important in cell-based assays designed to measure Gs- or Gi-coupled agonist stimulation or inhibition. Intra-assay precision for duplicate samples is typically 5% or less.
This competitive immunoassay is formatted with maximum flexibility to permit either manual assay or automated high-throughput screening. The cAMP immunoassay is based on the highly sensitive CSPD alkaline phosphate substrate, a chemiluminescent 1,2-dioxetane, with Sapphire-II luminescence enhancer. The ready-to-use substrate⁄enhancer reagent generates sustained glow light emission that is measured 30 minutes after addition. Once the substrate⁄ enhancer reaches the glow signal, the plate can be read for hours with little or no degradation of the signal, facilitating screening protocols in which several plates are compared to each other. In addition, the assay exhibits exceptionally low crossreactivity with other adenosine containing or cyclic nucleotides.
The cAMP Chemiluminescent Immunoassay Kit (2-plate size, C10557; 10-plate size, C10558) is designed for the rapid and sensitive quantitation of cAMP in extracts prepared from mammalian cells cultured in microwell plates. Each kit provides all required reagents, including:
- Alkaline phosphate conjugate of cAMP
- Anti-cAMP antibody
- cAMP standard
- CSPD substrate and Sapphire-II luminescence enhancer
- Assay and lysis buffer
- Conjugate dilution buffer
- Wash buffer
- Pre-coated microplates
- Detailed protocols (cAMP Chemiluminescent Immunoassay Kit)
The cAMP Chemiluminescent Immunoassay Kit is designed for quantitating cellular cAMP for functional assays of receptor activation. It has been used with established cell lines for functional measurements with endogenous receptors,![]() with cell lines containing exogenously expressed ligand receptors,
with cell lines containing exogenously expressed ligand receptors,![]() with primary cells
with primary cells ![]() and with tissues.
and with tissues.![]() It has also been used for receptor characterization,
It has also been used for receptor characterization,![]() orphan receptor ligand identification
orphan receptor ligand identification ![]() and the characterization of novel chimeric receptors.
and the characterization of novel chimeric receptors.![]() In addition, this assay can be used for high-throughput screening assays
In addition, this assay can be used for high-throughput screening assays ![]() of compounds that stimulate or interfere with these signal transduction pathways.
of compounds that stimulate or interfere with these signal transduction pathways.
Nucleotide analogs that serve as substrates or inhibitors of enzymes, as well as nucleotide derivatives that selectively bind to regulatory sites of nucleotide-binding proteins, have been used as structural and mechanistic probes for isolated proteins, reconstituted membrane-bound enzymes, organelles such as mitochondria, and tissues such as skinned muscle fibers.![]() More recently, however, these analogs have also been employed to study the effects of nucleotides on signal transduction and to screen for compounds that may affect signal transduction, such as G protein inhibitors and activators (G proteins and GTP Analogs for Binding Studies—Note 17.1).
More recently, however, these analogs have also been employed to study the effects of nucleotides on signal transduction and to screen for compounds that may affect signal transduction, such as G protein inhibitors and activators (G proteins and GTP Analogs for Binding Studies—Note 17.1).
We prepare a variety of nucleotide analogs, including:
- Alexa Fluor derivatives of cAMP for use as probes of type I cAMP-dependent protein kinases (PKA I) and Alexa Fluor 647 ATP (A22362)
- BODIPY dye–labeled nucleotides for use as enzyme substrates and as long-wavelength probes of nucleotide-binding sites
- Environment-sensitive, blue-fluorescent N-methylanthraniloyl (MANT) nucleotides
- Blue-fluorescent ethenoadenosine triphosphate (ε-ATP, E23691)
- Environment-sensitive trinitrophenyl (TNP) nucleotides
- Caged nucleotides, which are important probes for studying the kinetics and mechanism of nucleotide-binding proteins because they allow spatial and temporal control of the release of active nucleotide
- Photoaffinity nucleotides for site-selective covalent labeling
- Fluorescent ChromaTide nucleotides and aha-dUTP nucleotides, which are primarily used for biosynthetic incorporation into DNA or RNA (Labeling Oligonucleotides and Nucleic Acids—Section 8.2)
Alexa Fluor cAMP and Alexa Fluor ATP
Our Alexa Fluor cAMP analogs are 8-(6-aminohexyl)amino derivatives; similar analogs have been shown to exhibit a marked preference for binding to type I cAMP-dependent protein kinases (PKA I). We offer the green-fluorescent Alexa Fluor 488 cAMP (A35775) and far-red–fluorescent Alexa Fluor 647 cAMP (A35777). Alexa Fluor 488 cAMP was loaded into cells by electroporation and then used to measure intercellular diffusion of cAMP from regulatory to responder T cells via gap junctions.![]()
The Alexa Fluor 647 conjugate of ATP (A22362) comprises the long-wavelength Alexa Fluor 647 fluorophore linked to the ribose of ATP by a urethane bridge. Validated applications of this probe include fluorescence resonance energy transfer (FRET) analysis (Fluorescence Resonance Energy Transfer (FRET)—Note 1.2) of nucleotide association with Na+/K+-ATPase ![]() and measurements of the catalytic activity of heavy meromyosin.
and measurements of the catalytic activity of heavy meromyosin.![]()
BODIPY Ribonucleotide Di- and Triphosphates
Our selection of BODIPY dye–modified ribonucleotides includes:
- BODIPY FL adenosine 5'-triphosphate (BODIPY FL ATP, A12410)
- BODIPY TR adenosine 5'-triphosphate (BODIPY TR ATP, A22352)
- BODIPY TR adenosine 5'-diphosphate (BODIPY TR ADP, A22359)
- BODIPY FL guanosine 5'-triphosphate (BODIPY FL GTP, G12411)
- BODIPY TR guanosine 5'-triphosphate (BODIPY TR GTP, G22351)
- BODIPY FL guanosine 5'-diphosphate (BODIPY FL GDP, G22360)
These mixed-isomer analogs comprise a BODIPY fluorophore attached to the 2' or 3' position of the ribose ring via an aminoethylcarbamoyl linker. Interactions between the fluorophore and the purine base are evident from the spectroscopic properties of these nucleotide analogs. The fluorescence quantum yield of BODIPY FL GTP and BODIPY TR GTP is significantly quenched in solution (Figure 17.3.9) and increases upon binding to at least some GTP-binding proteins.![]() Similar nucleotide analogs incorporating fluorophores such as fluorescein, tetramethylrhodamine and Cy3 dye have been primarily used for biophysical studies of nucleotide-binding proteins.
Similar nucleotide analogs incorporating fluorophores such as fluorescein, tetramethylrhodamine and Cy3 dye have been primarily used for biophysical studies of nucleotide-binding proteins.![]() The BODIPY dye–labeled nucleotides may be particularly useful for fluorescence polarization–based assays of ATP- or GTP-binding proteins.
The BODIPY dye–labeled nucleotides may be particularly useful for fluorescence polarization–based assays of ATP- or GTP-binding proteins.

Figure 17.3.10 Principle of fluorescence-based detection of the diadenosine triphosphate hydrolase activity of Fhit using BODIPY FL GTP-γ-S thioester (G22183) as a substrate analog.
Nonhydrolyzable BODIPY ATP and GTP Analogs
Among the most useful fluorescent nucleotides for protein-binding studies are those that stoichiometrically bind to ATP- or GTP-binding sites but are not metabolized. We offer the following nonhydrolyzable BODIPY nucleotides:
- BODIPY FL AMPPNP
 (B22356)
(B22356) - BODIPY FL ATP-γ-S (A22184)
- BODIPY FL GTP-γ-S
 (G22183)
(G22183) - BODIPY 515/530 GTP-γ-S (G35779)
- BODIPY TR GTP-γ-S (G35780)
- BODIPY FL GTP-γ-NH amide (G35778)
The fluorescence of the BODIPY GTP-γ-S thioesters is quenched ~90% relative to that of the free dye but is recovered upon protein binding to G-proteins.![]() The green-fluorescent BODIPY FL GTP-γ-S has been used to detect GTP-binding proteins separated by capillary electrophoresis.
The green-fluorescent BODIPY FL GTP-γ-S has been used to detect GTP-binding proteins separated by capillary electrophoresis.![]() As compared with BODIPY FL GTP-γ-S thioester, the green-fluorescent BODIPY 515/530 GTP-γ-S thioester has a greater fluorescence increase upon protein binding, . The BODIPY TR GTP-γ-S thioester is a red-fluorescent analog with spectral properties similar to the Texas Red dye.
As compared with BODIPY FL GTP-γ-S thioester, the green-fluorescent BODIPY 515/530 GTP-γ-S thioester has a greater fluorescence increase upon protein binding, . The BODIPY TR GTP-γ-S thioester is a red-fluorescent analog with spectral properties similar to the Texas Red dye.
Although BODIPY FL GTP-γ-NH amide exhibits less fluorescence enhancement upon protein binding, it is reportedly the best of the three green-fluorescent GTP-γ analogs for directly monitoring nucleotide exchange.![]() The different linker lengths of the green-fluorescent GTP-γ analogs (six-carbon for BODIPY FL GTP-γ-NH amide, four-carbon for BODIPY FL GTP-γ-S and one-carbon for BODIPY 515/530 GTP-γ-S) may be useful for understanding protein active-site geometries.
The different linker lengths of the green-fluorescent GTP-γ analogs (six-carbon for BODIPY FL GTP-γ-NH amide, four-carbon for BODIPY FL GTP-γ-S and one-carbon for BODIPY 515/530 GTP-γ-S) may be useful for understanding protein active-site geometries.
In addition to their potential use for binding studies, BODIPY FL ATP-γ-S and BODIPY FL GTP-γ-S thioesters are important substrates for Fhit (Figure 17.3.10), a member of the histidine triad superfamily of nucleotide-binding proteins that bind and cleave diadenosine polyphosphates.![]() Fhit, one of the most frequently inactivated proteins in lung cancer, functions as a tumor suppressor by inducing apoptosis.
Fhit, one of the most frequently inactivated proteins in lung cancer, functions as a tumor suppressor by inducing apoptosis.![]() These BODIPY nucleotides should be especially useful for screening potential Fhit inhibitors and activators.
These BODIPY nucleotides should be especially useful for screening potential Fhit inhibitors and activators.

N-Methylanthraniloyl (MANT) Nucleotides
The blue-fluorescent MANT nucleotide analogs of ATP, AMPPNP, GTP, GMPPNP, ADP and GDP are modified on the ribose moiety, making these probes particularly useful for studying nucleotide-binding proteins that are sensitive to modifications of the purine base.![]() The compact nature of the MANT fluorophore and its attachment position on the ribose ring results in nucleotide analogs that induce minimal perturbation of nucleotide–protein interactions, as confirmed by X-ray crystal structures of MANT nucleotides bound to myosin
The compact nature of the MANT fluorophore and its attachment position on the ribose ring results in nucleotide analogs that induce minimal perturbation of nucleotide–protein interactions, as confirmed by X-ray crystal structures of MANT nucleotides bound to myosin ![]() and H-ras p21.
and H-ras p21.![]() Furthermore, because MANT fluorescence is sensitive to the environment of the fluorophore, nucleotide–protein interactions may be directly detectable. These properties (Spectroscopic properties of MANT-ATP in aqueous solution (pH 8)—Table 17.2) make MANT nucleotides valuable probes of the structure and enzymatic activity of nucleotide-binding proteins.
Furthermore, because MANT fluorescence is sensitive to the environment of the fluorophore, nucleotide–protein interactions may be directly detectable. These properties (Spectroscopic properties of MANT-ATP in aqueous solution (pH 8)—Table 17.2) make MANT nucleotides valuable probes of the structure and enzymatic activity of nucleotide-binding proteins.![]()
Applications for MANT-ATP (M12417), MANT-ADP (M12416) and MANT-AMPPNP (M22354) include analysis of:
- ATPase kinetics of kinesin
 and other microtubule motor-proteins
and other microtubule motor-proteins  using stopped-flow fluorescence measurements
using stopped-flow fluorescence measurements - Conformation of the myosin subfragment-1 nucleotide-binding site, as indicated by fluorescence quencher accessibility

- Interaction of P-glycoprotein ATP-binding sites with drug efflux–modulating steroids

- Myosin ATPase activity in rabbit skeletal muscle

- Structural characteristics of the nucleotide-binding site of Escherichia coli DnaB helicase

Applications for MANT-GTP (M12415), MANT-GDP (M12414) and MANT-GMPPNP (M22353) include analysis of:
- Activation of protein kinases by Rho subfamily GTP-binding proteins

- Conformational changes during activation of heterotrimeric G-proteins

- Effects of nucleotide structural modifications on binding to H-ras p21

- Nucleotide hydrolysis and dissociation kinetics of H-ras p21 and other low molecular weight GTP-binding proteins

- GTP-binding proteins Rab5 and Rab7,
 Raf-1,
Raf-1, Rho
Rho  and Rac,
and Rac, as well as Ras-related proteins
as well as Ras-related proteins
Ethenoadenosine Nucleotide
The ethenoadenosine nucleotides—developed in 1972 by Leonard and collaborators ![]() —bind like endogenous nucleotides to several proteins. The properties and applications of ethenoadenosine and MANT nucleotides have been comprehensively reviewed.
—bind like endogenous nucleotides to several proteins. The properties and applications of ethenoadenosine and MANT nucleotides have been comprehensively reviewed.![]() The etheno ATP analog (ε-ATP, E23691) can often mimic ATP in both binding and function. This probe has been used to replace ATP in actin polymerization reactions
The etheno ATP analog (ε-ATP, E23691) can often mimic ATP in both binding and function. This probe has been used to replace ATP in actin polymerization reactions ![]() and is frequently incorporated in place of the tightly bound actin nucleotide.
and is frequently incorporated in place of the tightly bound actin nucleotide.![]() It also supports contraction of actomyosin, facilitates the measurement of nucleotide-exchange kinetics in actin
It also supports contraction of actomyosin, facilitates the measurement of nucleotide-exchange kinetics in actin ![]() and serves as a substrate for myosin, which converts it to ε-ADP.
and serves as a substrate for myosin, which converts it to ε-ADP.![]() Sensitized luminescence of Tb3+ (T1247, Polar Tracers—Section 14.3) coordinated to ε-ATP is a sensitive probe of binding to the catalytic site of protein disulfide isomerase.
Sensitized luminescence of Tb3+ (T1247, Polar Tracers—Section 14.3) coordinated to ε-ATP is a sensitive probe of binding to the catalytic site of protein disulfide isomerase.![]()
Trinitrophenyl (TNP) Nucleotides
Unlike the etheno derivatives, the free trinitrophenyl (TNP) nucleotides are essentially nonfluorescent in water. The TNP nucleotides undergo an equilibrium transition to a semiquinoid structure that has relatively long-wavelength spectral properties;![]() this form is only fluorescent when bound to the nucleotide-binding site of some proteins. The TNP derivative of ATP frequently exhibits a spectral shift and fluorescence enhancement upon protein binding and actually binds with higher affinity than ATP to some proteins. The broad, long-wavelength absorption of TNP nucleotides makes them useful for FRET studies
this form is only fluorescent when bound to the nucleotide-binding site of some proteins. The TNP derivative of ATP frequently exhibits a spectral shift and fluorescence enhancement upon protein binding and actually binds with higher affinity than ATP to some proteins. The broad, long-wavelength absorption of TNP nucleotides makes them useful for FRET studies ![]() (Fluorescence Resonance Energy Transfer (FRET)—Note 1.2). The TNP derivatives of ATP (TNP-ATP, T7602), ADP (TNP-ADP, T7601) and AMP (TNP-AMP, T7624) have been used as structural probes for a wide variety of nucleotide-binding proteins.
(Fluorescence Resonance Energy Transfer (FRET)—Note 1.2). The TNP derivatives of ATP (TNP-ATP, T7602), ADP (TNP-ADP, T7601) and AMP (TNP-AMP, T7624) have been used as structural probes for a wide variety of nucleotide-binding proteins.![]() We have found that chromatographically purified TNP nucleotides are unstable during lyophilization. Consequently, these derivatives are sold in aqueous solution and should be frozen immediately upon arrival.
We have found that chromatographically purified TNP nucleotides are unstable during lyophilization. Consequently, these derivatives are sold in aqueous solution and should be frozen immediately upon arrival.
Caged Nucleotides
Caged nucleotides are nucleotide analogs in which the terminal phosphate is esterified with a blocking group, rendering the molecule biologically inactive. Photolytic removal of the caging group by UV illumination results in a pulse of the nucleotide—often on a microsecond to millisecond time scale—at the site of illumination. Because photolysis ("uncaging") can be temporally controlled and confined to the area of illumination, the popularity of this technique is growing. We are supporting this development by synthesizing a variety of caged nucleotides, neurotransmitters and Ca2+ chelators. Our current selection of caged nucleotides includes:
Photoactivatable Reagents, Including Photoreactive Crosslinkers and Caged Probes—Section 5.3 discusses our selection of caged probes and the properties of the different caging groups that we use (Properties of six different caging groups—Table 5.2).
Researchers investigating the cytoskeleton have benefited greatly from advances in caging technology, primarily originating from the work of Trentham, Kaplan and their colleagues.![]() NPE-caged ADP (A7056) is a useful probe for studying the effect of photolytic release of ADP in muscle fibers
NPE-caged ADP (A7056) is a useful probe for studying the effect of photolytic release of ADP in muscle fibers ![]() and isolated sarcoplasmic reticulum.
and isolated sarcoplasmic reticulum.![]() Although it is sometimes difficult to properly abstract papers that describe experiments with caged ATP because they could be referring to either NPE-caged ATP (A1048), DMNPE-caged ATP (A1049) or earlier caged versions of this nucleotide, most researchers have used NPE-caged ATP.
Although it is sometimes difficult to properly abstract papers that describe experiments with caged ATP because they could be referring to either NPE-caged ATP (A1048), DMNPE-caged ATP (A1049) or earlier caged versions of this nucleotide, most researchers have used NPE-caged ATP.
Because the caged nucleotides may be added to an experimental system at relatively high concentrations, use of the enzyme apyrase was recommended by Sleep and Burton ![]() to eliminate any traces of ATP that may be present in the caged ATP probes.
to eliminate any traces of ATP that may be present in the caged ATP probes.![]() Once the caged ATP solutions have been preincubated with apyrase, the enzyme can be removed by centrifugal filtration.
Once the caged ATP solutions have been preincubated with apyrase, the enzyme can be removed by centrifugal filtration.![]()
These caged nucleotides are generally cell impermeant and must be microinjected into cells or loaded by other techniques (Techniques for loading molecules into the cytoplasm—Table 14.1). Permeabilization of cells with staphylococcal α-toxin or the saponin ester β-escin is reported to make the membrane of smooth muscle cells permeable to low molecular weight (<1000 daltons) molecules, while retaining high molecular weight compounds.![]() α-Toxin permeabilization has permitted the introduction of caged nucleotides, including caged ATP (A1048) and caged GTP-γ-S, as well as of caged inositol 1,4,5-triphosphate (NPE-caged Ins 1,4,5-P3; I23580; Calcium Regulation—Section 17.2) into smooth muscle cells.
α-Toxin permeabilization has permitted the introduction of caged nucleotides, including caged ATP (A1048) and caged GTP-γ-S, as well as of caged inositol 1,4,5-triphosphate (NPE-caged Ins 1,4,5-P3; I23580; Calcium Regulation—Section 17.2) into smooth muscle cells.![]() Caged inositol 1,4,5-triphosphate has also been successfully loaded in ECV304 cells using electroporation.
Caged inositol 1,4,5-triphosphate has also been successfully loaded in ECV304 cells using electroporation.![]()
BzBzATP
Functional ion channels can be assembled from both homomeric and heteromeric combinations of the seven P2X receptor subunits so far identified (P2X1–7). Due to the lack of specific agonists or antagonists for P2X receptors, it is difficult to determine which receptor subtypes mediate particular cellular responses. We offer one of the most potent and widely used P2X receptor agonists, BzBzATP ![]() (2'-(or 3'-)O-(4-benzoylbenzoyl)adenosine 5'-triphosphate, B22358). BzBzATP has more general applications for site-directed irreversible modification of nucleotide-binding proteins via photoaffinity labeling.
(2'-(or 3'-)O-(4-benzoylbenzoyl)adenosine 5'-triphosphate, B22358). BzBzATP has more general applications for site-directed irreversible modification of nucleotide-binding proteins via photoaffinity labeling.![]()
| Cat # | MW | Storage | Soluble | Abs | EC | Em | Solvent | Notes |
|---|---|---|---|---|---|---|---|---|
| A1048 | 700.30 | FF,D,LL | H2O | 259 | 18,000 | none | MeOH | 1, 2, 3 |
| A1049 | 760.35 | FF,D,LL | H2O | 351 | 4400 | none | H2O | 1, 2 |
| A7056 | 614.44 | FF,D,LL | H2O | 259 | 15,000 | none | MeOH | 1, 2, 3 |
| A12410 | 933.30 | FF,L | H2O | 505 | 54,000 | 514 | H2O | 4, 5 |
| A12412 | 1117.18 | FF,L | H2O | 323 | 4200 | 461 | pH 7 | 4, 5, 6 |
| A22184 | 878.28 | FF,L | H2O | 504 | 68,000 | 514 | pH 7 | 4, 5 |
| A22352 | 1065.43 | FF,L | H2O | 591 | 55,000 | 620 | pH 7 | 4, 5 |
| A22359 | 963.47 | FF,L | H2O | 592 | 57,000 | 621 | pH 7 | 4, 5 |
| A22362 | ~2050 | FF,L | H2O | 648 | 246,000 | 667 | pH 7 | 4, 5 |
| A35775 | 1162.23 | FF,L | H2O | 493 | 71,000 | 517 | pH 7 | 4, 5 |
| A35777 | ~1700 | FF,L | H2O | 649 | 246,000 | 666 | pH 7 | 4, 5 |
| B7469 | 784.70 | F,D,L | DMSO | 504 | 79,000 | 511 | MeOH | |
| B22356 | 932.31 | FF,L | H2O | 504 | 68,000 | 514 | H2O | 4, 5 |
| B22358 | 1018.97 | FF,L | H2O | 260 | 27,000 | none | pH 7 | |
| D1037 | 524.38 | F,D,LL | DMSO | 338 | 6100 | none | MeOH | 1, 2 |
| E23691 | 619.13 | FF | H2O | 265 | 5000 | 411 | pH 7 | 5 |
| G12411 | 949.30 | FF,L | H2O | 504 | 68,000 | 511 | H2O | 4, 5, 7 |
| G22183 | 894.28 | FF,L | H2O | 504 | 68,000 | 510 | pH 7 | 4, 5, 7 |
| G22351 | 1081.43 | FF,L | H2O | 591 | 56,000 | 620 | pH 7 | 4, 5, 7 |
| G22360 | 1005.75 | FF,L | H2O | 504 | 68,000 | 508 | pH 7 | 4, 5, 7 |
| G35778 | 905.29 | FF,L | H2O | 505 | 68,000 | 512 | pH 7 | 4, 5, 7 |
| G35779 | 865.28 | FF,L | H2O | 511 | 520 | pH 7 | 4, 5, 7 | |
| G35780 | 1153.60 | FF,L | H2O | 591 | 621 | pH 7 | 4, 5, 7 | |
| H7476 | 504.45 | F,D,L | DMSO, DMF | 591 | 37,000 | 594 | EtOH | |
| M12414 | 620.32 | FF,L | H2O | 356 | 5700 | 447 | pH 8 | 4, 5, 8 |
| M12415 | 722.28 | FF,L | H2O | 356 | 5700 | 448 | pH 7 | 4, 5, 8 |
| M12416 | 604.32 | FF,L | H2O | 356 | 5800 | 448 | pH 7 | 4, 5, 8 |
| M12417 | 706.28 | FF,L | H2O | 356 | 5800 | 447 | pH 7 | 4, 5, 8 |
| M22353 | 721.29 | FF,L | H2O | 357 | 5700 | 447 | pH 8 | 4, 5, 8 |
| M22354 | 705.29 | FF,L | H2O | 357 | 5800 | 447 | pH 8 | 4, 5, 8 |
| T7601 | 682.26 | FF,L | H2O | 408 | 26,000 | none | pH 8 | 4, 5, 9 |
| T7602 | 784.22 | FF,L | H2O | 408 | 26,000 | none | pH 8 | 4, 5, 9 |
| T7624 | 579.29 | F,L | H2O | 408 | 26,000 | none | pH 8 | 4, 5, 9 |
| ||||||||
For Research Use Only. Not for use in diagnostic procedures.
