I currently use Sanger sequencing
Many researchers who perform Sanger sequencing by capillary electrophoresis are now incorporating next-generation sequencing (NGS) techniques into their research, especially when higher throughput sequencing is desired, as well as RNA sequencing and sequencing of panels. Next-generation sequencing is opening doors to new studies that could not be accomplished in a practical manner with other sequencing technologies.
If you are considering or currently using Sanger sequencing for applications where many targets per sample are being analyzed, these applications may be good candidates for NGS, especially when suitable Ion AmpliSeq Ready-to-Use Panels are available. Exome sequencing and transcriptome sequencing applications can also be accomplished efficiently using NGS, due to the higher throughput capacity. Many of these applications can be performed in a fraction of the time and at a fraction of the cost of Sanger sequencing.
Sanger sequencing is recommended for confirmation and verification of NGS results, and products such as the Sanger sequencing online Primer Designer Tool enable you to quickly perform confirmation studies. Select from >300,000 predesigned primer pairs targeting the human exome using this tool.
It is also typically most practical to continue using Sanger sequencing for DNA sequencing applications such as plasmid sequencing to check that the correct DNA was inserted into a vector. Sanger sequencing remains the gold standard sequencing technology, so any applications where base-calling accuracy is critical should use Sanger sequencing for confirmation, at least until the accuracy of the NGS technique is verified. Sanger sequencing is also useful where long reads (>1,000 bases) are required and for sequencing through difficult regions that may not yield results with NGS. Finally, the capillary electrophoresis techniques employed in Sanger sequencing continue to be useful in fragment analysis applications, such as microsatellite analysis.
NGS and Sanger sequencing use many of the same standard molecular biology techniques to prepare samples for sequencing. Sanger sequencing requires amplification of the target of interest and a cycle sequencing reaction prior to analysis by capillary electrophoresis, whereas NGS requires library preparation and template preparation prior to the sequencing reaction.
Isolation of DNA from samples of interest, a critical first step for any Sanger sequencing application.
Amplify the DNA region of interest: select from predesigned primer pairs or design your own.
Generate sequencing products by cycle sequencing; purify reactions to remove fluorescent ddNTP.
The products of the sequencing reaction are injected into capillaries filled with polymer to perform capillary electrophoresis.
Translate raw data into corresponding electropherograms using downstream software applications.
Select from our pre-designed gene panels or design your own with Ion AmpliSeq™ Designer.
Choose the Ion OneTouch™ 2 System or the Ion Chef ™ System* for fully automated template preparation.
The Ion PGM™ and Ion Proton™ Sequencers can handle any application and are priced for any lab.
Torrent Suite™ Software provides automated data analysis and flows into application-specific workflows for biological interpretation.
Getting started with next-generation sequencing
Since the massively parallel "sequencing by synthesis" approach for next-generation sequencing is different from the chain-termination method used in Sanger sequencing, some work is required to directly transition Sanger sequencing assays to NGS. In many cases, labs continue to use Sanger sequencing for their lower throughput applications and embark upon new studies using NGS.
Transitioning assays to next-generation sequencing
An easy way to transition from Sanger sequencing assays to NGS is through the use of ready-to-use panels such as the Ion AmpliSeq panels. This helps you avoid the need to design new primers for NGS and greatly simplifies the library construction step, enabling sequencing to be completed in a single day. Custom panels can also be designed in about 2 hours.
Researchers studying genetic variation have a simple, fast, and affordable solution for sequencing specific human genes or genomic regions: Ion AmpliSeq targeted selection technology. This technology, in combination with the Ion PGM Sequencer, helps fast-track projects and allows researchers to expand their research by providing the flexibility to analyze hundreds of genes simultaneously.
Overview of startup and running costs for next-generation sequencing
The startup costs for next-generation sequencing have become quite affordable with the introduction of benchtop NGS systems. Benchtop systems can typically be purchased for $100,000–$150,000. The most affordable is the Ion PGM Sequencer, which is about half the cost of other benchtop systems. Running costs range from as little as $100–$200 per sample when samples are multiplexed, to ~$350–$1,000 per sample without multiplexing. Per-target costs are much lower for NGS since the capacity is so much higher than that of Sanger sequencing; thousands of targets per sample can be analyzed in a single NGS run, so the per-target cost can be as low as a few cents.
Data analysis—overcoming bottlenecks
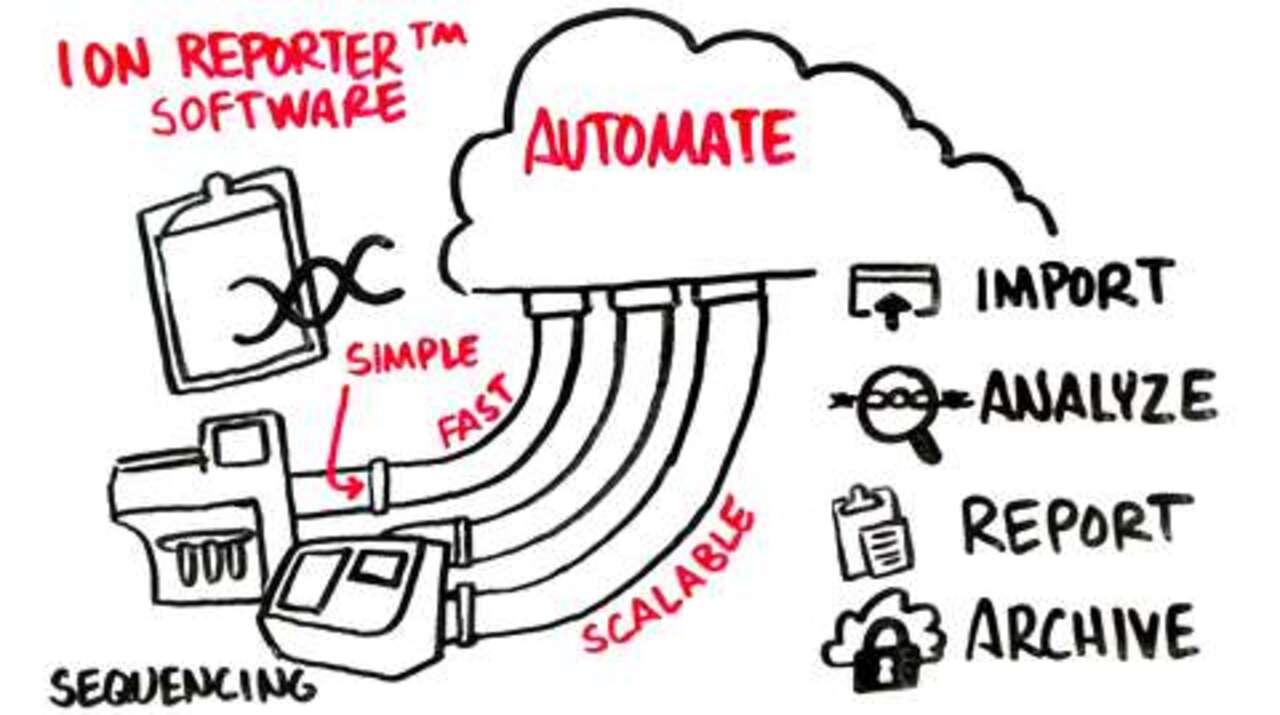
One of the most significant adjustments in transitioning to next-generation sequencing is the need for dedicated bioinformatics resources to handle the massive amounts of data generated by NGS platforms. While the readouts from Sanger sequencing are easy-to-interpret chromatograms that display the nucleotide at each position and then mapped to your reference sequence, the readouts from NGS have ten- to many hundred-fold read redundancy (to derive an equivalent consensus read at high accuracy) and thus require secondary analysis. Ion Torrent next-generation sequencing has helped overcome this bottleneck by integrating informatics solutions such as Ion Reporter Software, which allows you to do the read data processing, read mapping, variant calling, and variant filtering to interpret your data quickly and unambiguously.
Ion Reporter Software comprises a suite of bioinformatics tools that streamline and simplify analysis, annotation, and archiving of Ion semiconductor sequencing data.
Designed specifically for researchers performing routine sequencing assays, Ion Reporter Software helps you overcome the bioinformatics barriers and interpret DNA variants faster and more consistently.
Resources
Support
NewCapillary Electrophoresis Instruments Support Center
Find tips, troubleshooting help, and resources for support related to your capillary electrophoresis instruments.
| NEW | Next-Generation Sequencing Support Center Find tips, troubleshooting help, and resources for your next-gen sequencing applications. |
For Research Use Only. Not for use in diagnostic procedures.
For Research Use Only. Not for use in diagnostic procedures.