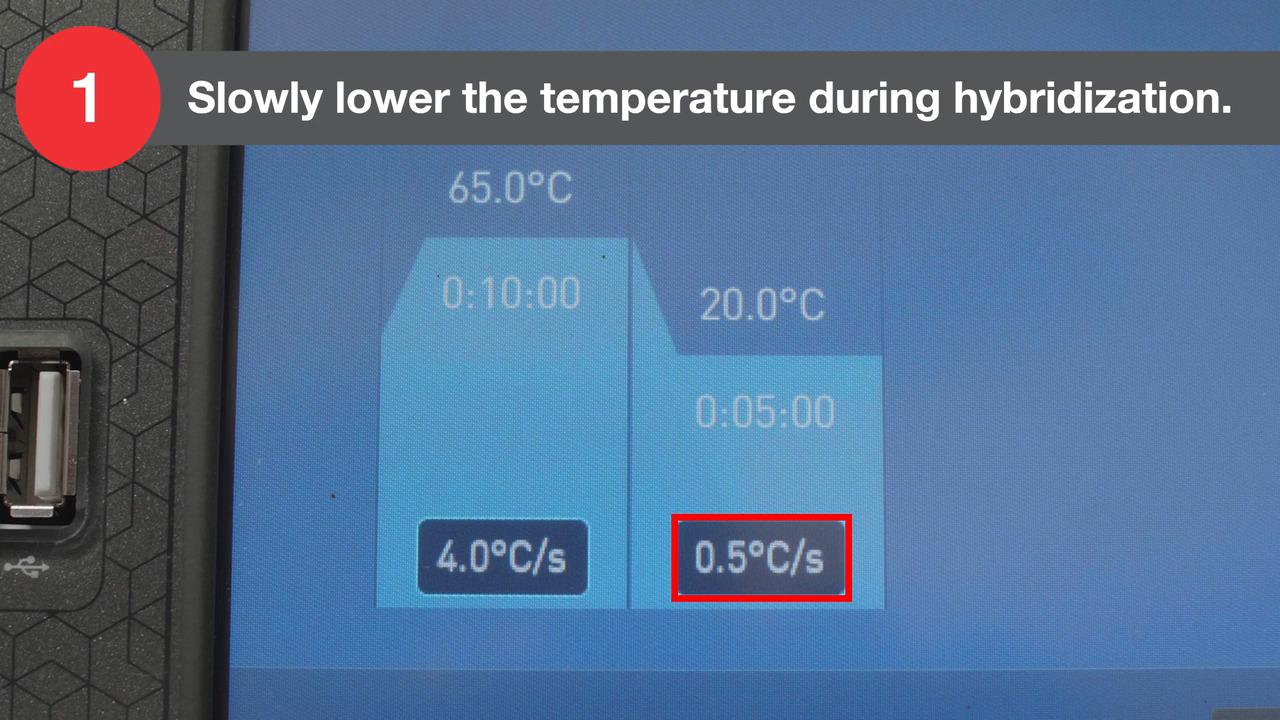
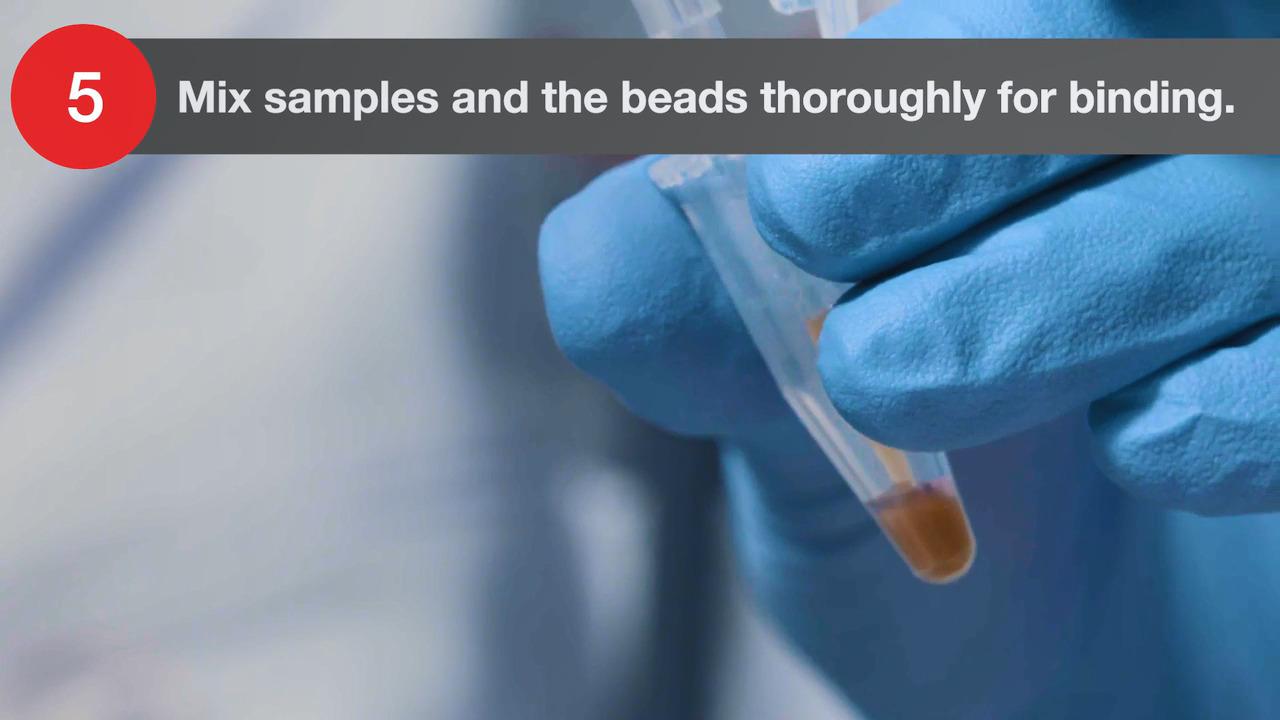
Reveal the full complexity of a plant transcriptome or perform bacterial RNA sequencing using the Invitrogen Collibri Stranded RNA Library Prep kits for Illumina systems.
- Suitable for bacterial RNA sequencing studies or plant transcriptome studies
- <4.5 hours for library generation following ribosomal RNA (rRNA) depletion
- For highest success rates, tracking dyes in each reagent change color when properly mixed
Library preparation for bacterial RNA sequencing or plant transcriptome studies is a two-step process.
Step 1. Deplete ribosomal RNA
The Invitrogen RiboMinus Isolation kits enable efficient and selective removal of ribosomal RNA (rRNA) from intact total RNA in less than two hours. Depleted RNA produced by RiboMinus kits is compatible with Collibri Library Prep kits in step two.
 RiboMinus Bacteria 2.0 Transcriptome Isolation Kit |  RiboMinus Plant Kit for RNA-Seq | |
| Species compatibility | Pan-prokaryote depletion probes have been shown to be compatible with over 76 species. | Wheat, maize, rice, barley, soybean, sugar cane, sorghum, potato, cassava, Arabidopsis, tomato, tobacco, poplar, legume, and moss |
| Sample quality compatibility | RIN values from 5.0–10.0 | RIN values from 8.0–10.0 |
| Hands-on time | 0.75 hours | 1 hour |
| Total time | <2 hours | 2 hours |
| rRNA depleted | 16S 23S 5S | nuclear (25/26S and 17/18S) chloroplast (23S and 16S) mitochondrion (18S) |
| Probe design | 150 probes targeting conserved regions of rRNA across 76 pan-prokaryote species | Probe mixture contains 6 probes. Each probe is specific for one or more of the conserved regions of the large cytosolic and chloroplast rRNA transcripts |
| Input range | 100 ng to 5 µg of total RNA | 1–10 µg total RNA |
| Download manual | Download manual |
Step 2. Rapid library generation
 Collibri Stranded RNA Library Prep Kit for Illumina Systems | |
| Hands-on time | 1.5 hours |
| Total time | 4.5 hours |
| RNA source | RNA depleted of ribosomal RNA (rRNA) |
| Input range | 1–25 ng of mRNA |
| Species compatibility | All |
| Ribosomal depletion module | Available separately |
| Analysis difficulty | Biologist-friendly |
The Collibri Stranded RNA Library Prep kits for Illumina systems generate NGS libraries in 4.5 hours and maintain sample complexity. The kits use a unique protocol in which partial adaptors, also known as helper adapters, are ligated directly to single-stranded RNA prior to creation of cDNA. Full-length indexed adaptors are generated during PCR. Collibri Stranded RNA Library Prep kits are compatible with all RNA samples following ribosomal depletion.
Technical Note: How strandedness is maintained during library preparation
Detect which strand of the genome encodes the gene transcript and differentiate between two distinct alleles encoded on opposite strands of the genome.
Visual feedback at each step in the workflow
For highest success rate, tracking dyes change color at each step only when proper mixing is achieved. Lack of a color change indicates the need to pause and complete mixing prior to continuing. Visual feedback is useful to monitor the performance of robotic systems.
Video: Collibri NGS Library Prep kits for Illumina systems
Tips for your RNA-seq library preparation
The scientists who developed the Collibri Stranded RNA Library Prep kits share their tips for cDNA library generation which retains the full transcriptome.
Recommended controls
ERCC RNA Spike-In Mixes are recommended to be added to the input RNA before library preparation. The mixes (not included in the kit) provide a set of external RNA controls that enable performance assessment for dynamic range, lower limit of detection, and fold-change response for gene expression experiments. For detailed information, refer to the ERCC RNA Spike-In Control Mixes User Guide.
For positive RNA control, we recommend using Universal Human Reference RNA (Cat. No. QS0639) or Human Brain Total RNA (Cat. No. AM7962).
Tip!
Do not use the ERCC RNA Spike-In Control Mixes with highly degraded samples such as FFPE RNA. High quality ERCC RNA can outcompete lower quality RNA population thus causing ERCC to be overrepresented in the final library.
Whole-transcriptome library preparation workflow
Retain coding and noncoding RNA diversity
We offer a range of Invitrogen genomic RNA extraction kits for sensitive, scalable purification from an expansive set of starting materials to help maximize process efficiency and downstream performance. This includes a broad range of kits for purifying RNA from a variety of samples including tissue, cells, blood, serum, plants, forensic samples, and more.
Automate RNA extraction using magnetic bead-based technology and KingFisher instruments to bind, wash, and elute RNA to reduce your hands-on time while maintaining high yields and excellent reproducibility.
Sensitive and specific nucleic acid quantification
Accurately and quickly quantify RNA with high sensitivity and specificity with the Qubit fluorometric quantification platforms. Choose from broad range or high sensitivity RNA quantification reagents to quantify RNA library preps.
Ribosomal depletion for bacteria or plants
The RiboMinus Isolation kits enable efficient and selective removal of ribosomal RNA (rRNA) from intact total RNA in less than two hours and are compatible with Collibri Library Prep kits.
Recommended accessories
- Interchange the 96-well 0.2-mL block with another block type if your throughput needs change
- Access the system remotely (and conveniently) through a mobile app
- Program the instrument in seconds with a simple-to-use touchscreen interface
- VeriFlex blocks allow for precise PCR optimization or to run up to six independent assays
- Easy-to-use graphical interface (6.5-inch VGA Touch Screen)
- Reduced PCR cycling time when using faster ramp rates
Quantification options to scale throughput
The qPCR-based Collibri Library Quantification kit scales well for larger sample batches and is the ideal method for precious samples or clinical samples. Quantification accuracy is equivalent to the KAPA Library Quantification kit with the additional benefit of visual feedback during the quantification process.
The Qubit dsDNA HS Assay is a fluorometric assay that uses dsDNA-binding dyes in order to accurately determine NGS library concentration, and benefits from a simple workflow of just a few minutes per sample.
Regardless of the assay that is chosen, good laboratory technique should be used in order to ensure accurate measurement of library concentrations and high-quality Illumina sequencing data.
Recommended accessories
With Applied Biosystems QuantStudio real-time PCR systems, you get true value with excellent performance, reliability, and world-class support. Our family of instruments enables you to obtain the results you need, connect and collaborate with colleagues, and achieve your research goals.
Retain coding and noncoding RNA diversity
We offer a range of Invitrogen genomic RNA extraction kits for sensitive, scalable purification from an expansive set of starting materials to help maximize process efficiency and downstream performance. This includes a broad range of kits for purifying RNA from a variety of samples including tissue, cells, blood, serum, plants, forensic samples, and more.
Automate RNA extraction using magnetic bead-based technology and KingFisher instruments to bind, wash, and elute RNA to reduce your hands-on time while maintaining high yields and excellent reproducibility.
Sensitive and specific nucleic acid quantification
Accurately and quickly quantify RNA with high sensitivity and specificity with the Qubit fluorometric quantification platforms. Choose from broad range or high sensitivity RNA quantification reagents to quantify RNA library preps.
Ribosomal depletion for bacteria or plants
The RiboMinus Isolation kits enable efficient and selective removal of ribosomal RNA (rRNA) from intact total RNA in less than two hours and are compatible with Collibri Library Prep kits.
Recommended accessories
- Interchange the 96-well 0.2-mL block with another block type if your throughput needs change
- Access the system remotely (and conveniently) through a mobile app
- Program the instrument in seconds with a simple-to-use touchscreen interface
- VeriFlex blocks allow for precise PCR optimization or to run up to six independent assays
- Easy-to-use graphical interface (6.5-inch VGA Touch Screen)
- Reduced PCR cycling time when using faster ramp rates
Quantification options to scale throughput
The qPCR-based Collibri Library Quantification kit scales well for larger sample batches and is the ideal method for precious samples or clinical samples. Quantification accuracy is equivalent to the KAPA Library Quantification kit with the additional benefit of visual feedback during the quantification process.
The Qubit dsDNA HS Assay is a fluorometric assay that uses dsDNA-binding dyes in order to accurately determine NGS library concentration, and benefits from a simple workflow of just a few minutes per sample.
Regardless of the assay that is chosen, good laboratory technique should be used in order to ensure accurate measurement of library concentrations and high-quality Illumina sequencing data.
Recommended accessories
With Applied Biosystems QuantStudio real-time PCR systems, you get true value with excellent performance, reliability, and world-class support. Our family of instruments enables you to obtain the results you need, connect and collaborate with colleagues, and achieve your research goals.
Ordering information
Resources
App note
Brochures
DNA sequencing
Technical notes
All sequencing methods
Technical notes
RNA sequencing
Technical notes
For Research Use Only. Not for use in diagnostic procedures.