Why use serial block-face imaging for volume electron microscopy?
Unraveling the complex 3D architecture of cells and tissues in their natural context is crucial for the structure-function correlation in biological systems. In recent years, there have been considerable advances in SEM-based methods for 3D reconstruction of large tissue volumes. Serial block-face SEM (SBF-SEM) combines in situ sectioning and imaging of plastic embedded tissue blocks within the SEM vacuum chamber in a fully automated fashion for reconstruction of large tissue volumes. Until now, the axial resolution was limited by the minimal section thickness that can be cut from the block-face; however, with a combination of SBF-SEM and multi-energy deconvolution SEM (MED-SEM), the Thermo Scientific Volumescope 2 SEM now enables large-volume imaging with truly isotropic 3D resolution.
How does serial block-face microscopy work?
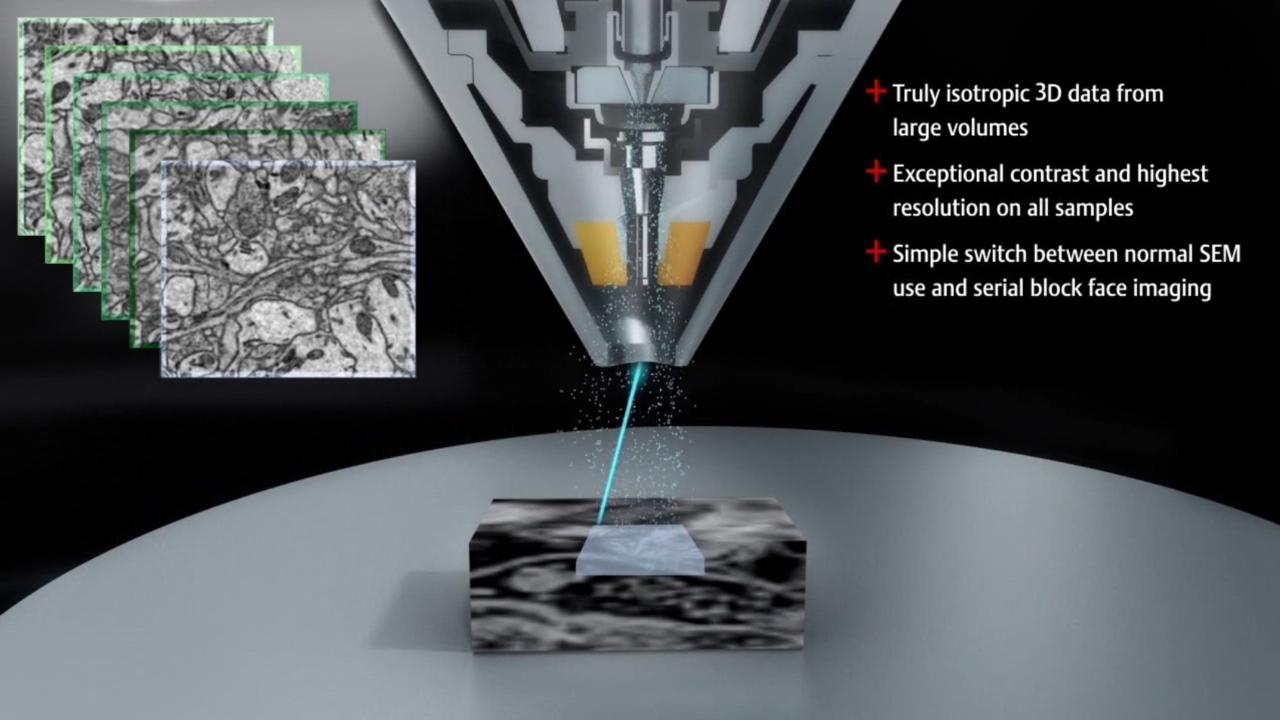
During serial block-face imaging, the electron beam is first used to scan the surface of a resin-embedded tissue sample, capturing a 2D image of the specimen.
This top surface is subsequently removed with an in situ microtome. The thickness of each section is user-defined but is typically greater than 15-20 µm. Once this section is discarded, it is gathered by a debris collection device.
An image of the fresh surface is then collected with the SEM. This process is repeated until the whole sample has been imaged; total sample height can range from tens to hundreds of micrometers or more. The serial stack of images is then processed using 3D rendering software.
The serial block-face imaging process can be optimized and refined to match specific user or sample needs and requirements, including localized regions of interest, multiple areas, or various imaging detectors.
Multi-energy deconvolution SEM (MED-SEM) is a novel way to improve the axial resolution by combining mechanical sectioning with optical sectioning. Following in situ sectioning of the block-face using a diamond knife, the freshly exposed tissue is imaged several times using increasing accelerating voltages. These images are subsequently used in a deconvolution algorithm to derive several optical subsurface layers, forming a 3D subset. By repeating this cycle, the Volumescope 2 SEM offers isotropic datasets with 10 nm z-resolution.
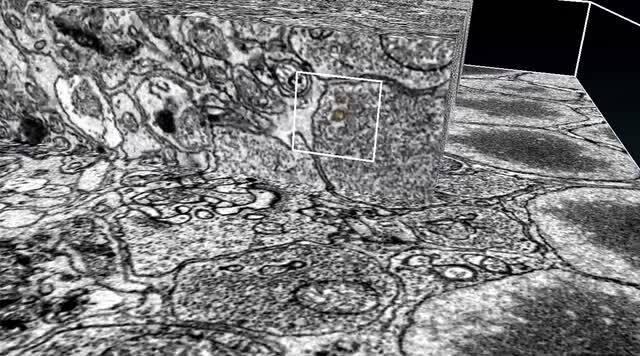
Serial block-face SEM 3D sample gallery
Mitochondria ultrastructure
Standardized protocol using the visualization software, Thermo Scientific Amira, to quantify organelle morphologies in 3D, thereby providing accurate and reproducible measurements of these cellular substructures. Protocol was applied with SBF-SEM and Amira software to quantify mitochondria and endoplasmic reticulum (ER) structures.

Meet the scientists using serial block-face scanning electron microscopy
“I used to surf the web. Now I surf datasets. Structure is function and that is the dogma of cell biology.”
Prof. Jeffrey Salisbury, Mayo Clinic, discusses his over 45 years of microscopy experience to answer various biological questions including recent 3D investigations with serial block-face imaging to look at 3D architecture of various specimens from tissue, kidney, and brain in normal and diseased states.
“It’s incredibly exciting times and finally we have an opportunity to do some real cell biology and for all these years I have been sitting and waiting for and I can’t wait to get on with it.”
Christopher Gilpin, Director of the Purdue Electron Microscopy Center, on discussing serial block-face imaging and its potential use for biological multi-scale mapping within the context of 3D imaging techniques.
Serial block-face imaging resources
For Research Use Only. Not for use in diagnostic procedures.